Microbial growth and death
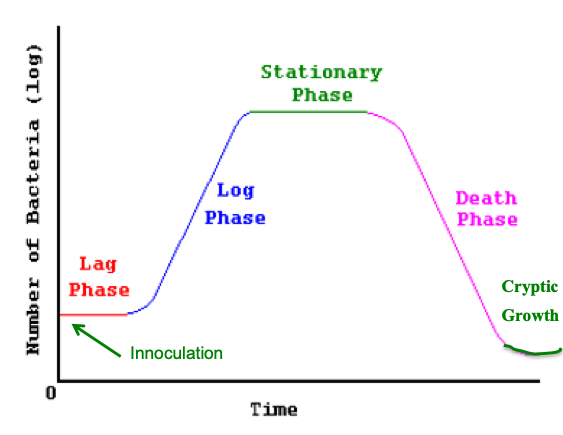
The growth cycle of microorganisms includes 5 distinct phases, each of which can be modeled with emprical and causal models (Chapter 5 of the textbook).
• lag phase
• log (exponential) phase
• stationary phase
•. death phase
• cryptic growth
These models and there implications are described in this lecture.
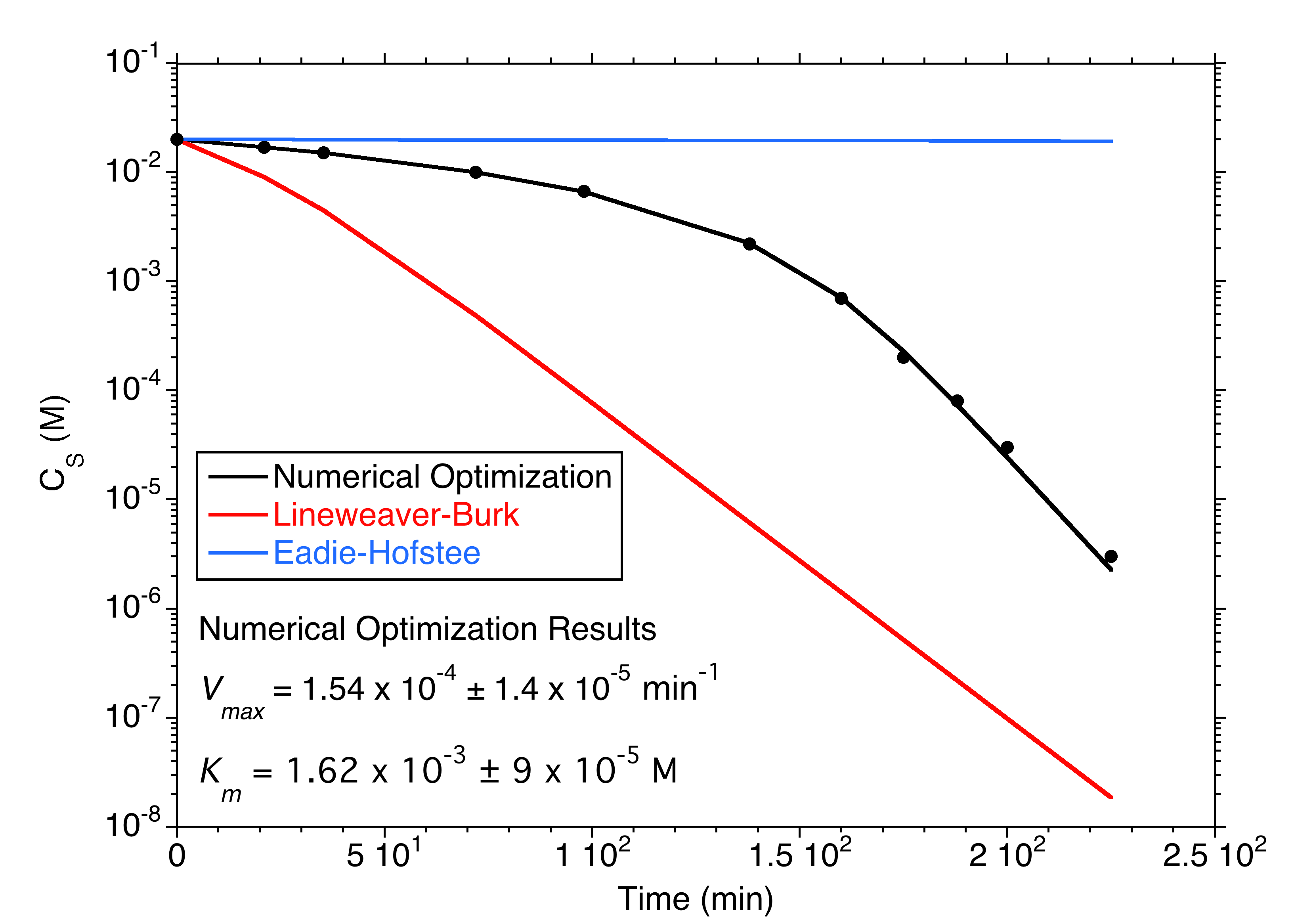
The proper way to fit Michaelis-Menten kinetic data
Traditional linearizations (Lineweaver-Burke and Eadie-Hoftsee) of the Michaelis-Menten equation to obtain Vmax and Km values from enzyme kinetic data can generate huge errors in these estimates. With the advent of modern computers it is easy to set up an implicit non-linear curve fitting routine in any spreadsheet using numerical optimization to accomplish this task both to get improved estimates and to establish the statistical confidence intervals in those estimates. This lecture (Chapter 6 of the textbook) describes how this is done. There is an accompanying spreadsheet demonstrating this in the Book Resources.
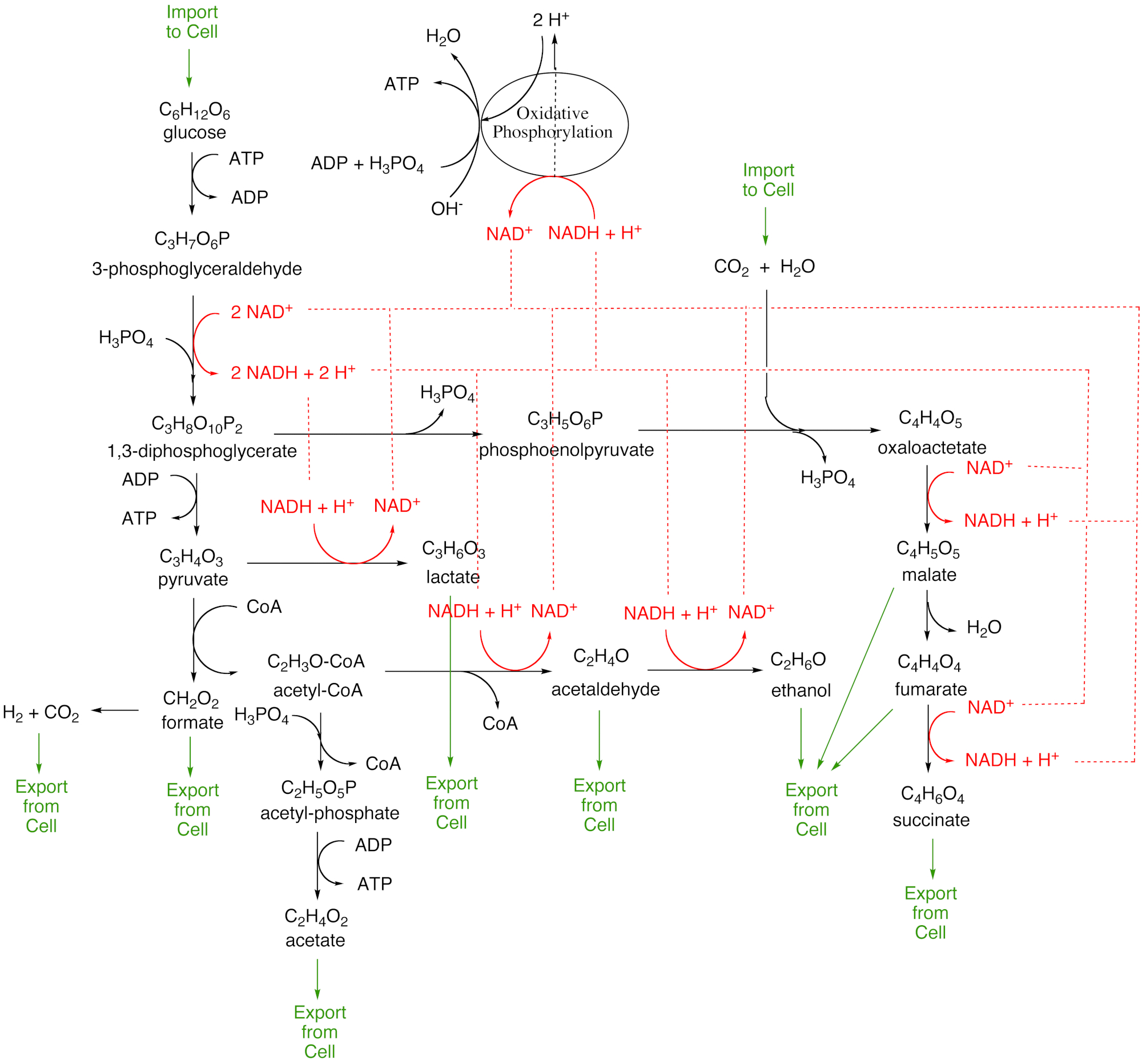
Metabolic uncoupling is life’s secret to adaptation and evolution
The capture of electrons and phosphate by biochemical intermediates is what enables metabolic networks to adapt and evolve. It is also the resaon that we can never obtain an exact stoichiometry for metabolic networks. In this lecture (taken from Chapter 2 of the textbook) we show why.
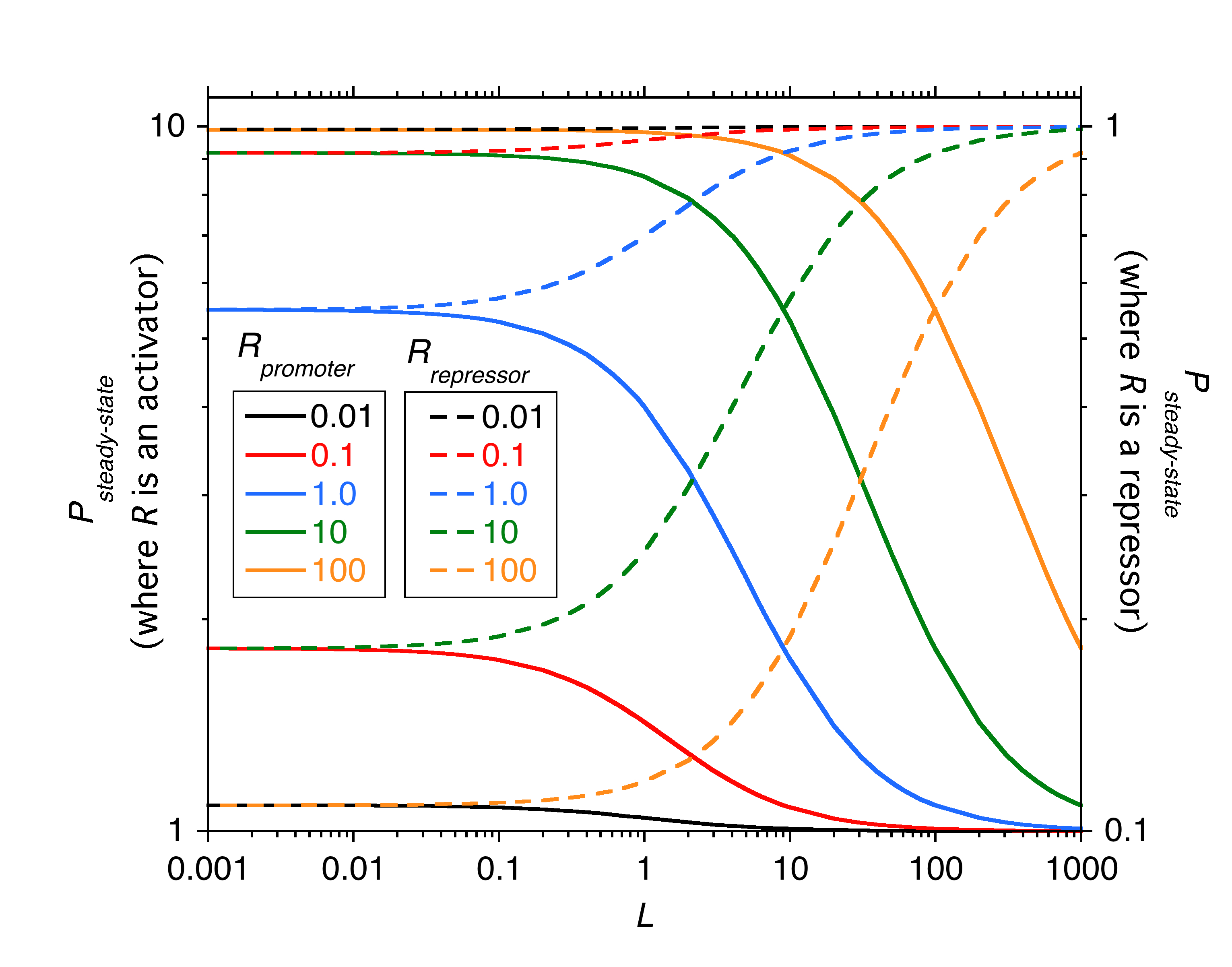
Transcriptional regulation
We now know that the expression of virtually all structural genes are tightly regulated, many by multiple regulatory mechanisms. In this lecture (taken from Chapter 10 of the textbook) we explore how this regulation occurs can be modeled. Chapters 10 through 13 of the textbook expand upon this methodology to model the lac and hut operons, and the pho regulon.
(C) 2021, LVS Sciences, LLC. All rights reserved.
LVS Sciences, LLC
1321 Upland Dr. Suite
Houston, TX 77043
United States of America.